This year’s Lasker Basic Medical Research Award was awarded to Demis Hassabis and John Jumper for their contributions to the creation of the AlphaFold artificial intelligence system that predicts the three-dimensional structure of proteins based on the first order sequence of amino acids.
Their results solve a problem that has long vexed the scientific community and open the door to accelerating research across the biomedical field. Proteins play a pivotal role in disease development: in Alzheimer’s disease, they fold and clump together; In cancer, their regulatory function is lost; In inborn metabolic disorders, they are dysfunctional; In cystic fibrosis, they go into the wrong space in the cell. These are just a few of the many mechanisms that cause disease. Detailed protein structure models can provide atomic configurations, drive the design or selection of high-affinity molecules, and accelerate drug discovery.
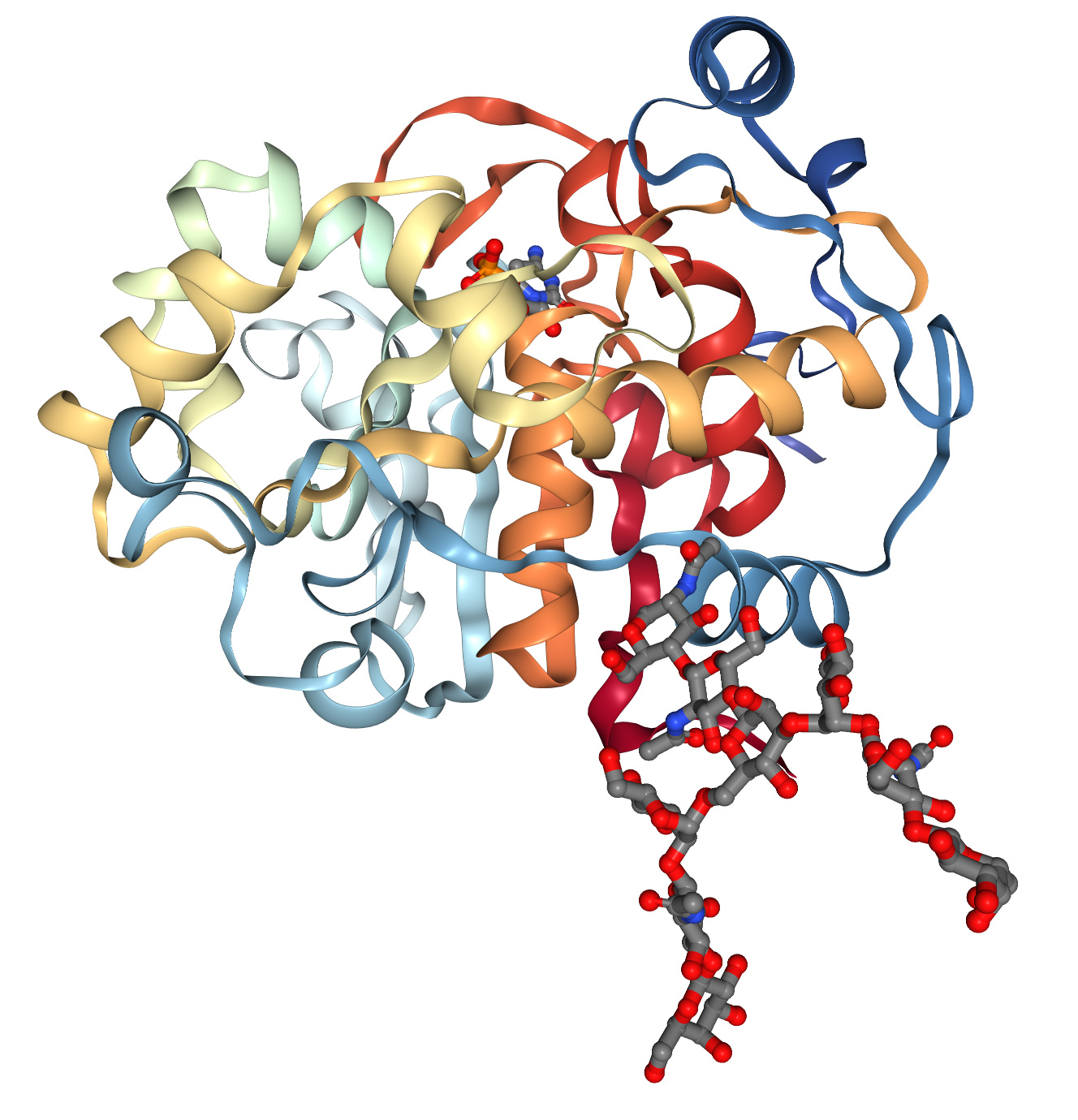
Protein structures are generally determined by X-ray crystallography, nuclear magnetic resonance and cryo-electron microscopy. These methods are expensive and time-consuming. This results in existing 3D protein structure databases with only about 200,000 structural data, while DNA sequencing technology has produced more than 8 million protein sequences. In the 1960s, Anfinsen et al. discovered that the 1D sequence of amino acids can spontaneously and repeatably fold into a functional three-dimensional conformation (Figure 1A), and that molecular “chaperones” can accelerate and facilitate this process. These observations lead to a 60-year challenge in molecular biology: predicting the 3D structure of proteins from the 1D sequence of amino acids. With the success of the Human Genome Project, our ability to obtain 1D amino acid sequences has greatly improved, and this challenge has become even more urgent.
Predicting protein structures is difficult for several reasons. First, all possible three-dimensional positions of every atom in every amino acid require a lot of exploration. Second, proteins make maximum use of complementarity in their chemical structure to efficiently configure atoms. Since proteins typically have hundreds of hydrogen bond “donors” (usually oxygen) that should be close to the hydrogen bond “acceptor” (usually nitrogen bound to hydrogen), it can be very difficult to find conformations where nearly every donor is close to the acceptor. Third, there are limited examples for the training of experimental methods, so it is necessary to understand the potential three-dimensional interactions between amino acids on the basis of 1D sequences using information on the evolution of the relevant proteins.
Physics was first used to model the interaction of atoms in the search for the best conformation, and a method was developed to predict the structure of proteins. Karplus, Levitt and Warshel were awarded the 2013 Nobel Prize in Chemistry for their work on computational simulation of proteins. However, physics-based methods are computationally expensive and require approximate processing, so precise three-dimensional structures cannot be predicted. Another “knowledge-based” approach is to use databases of known structures and sequences to train models through artificial intelligence and machine learning (AI-ML). Hassabis and Jumper apply elements of both physics and AI-ML, but the innovation and leap in performance of the approach stems primarily from AI-ML. The two researchers creatively combined large public databases with industrial-grade computing resources to create AlphaFold.
How do we know they have “solved” the structural prediction puzzle? In 1994, the Critical Assessment of Structure Prediction (CASP) competition was established, which meets every two years to track the progress of structural prediction. The researchers will share the 1D sequence of the protein whose structure they have recently resolved, but whose results have not yet been published. The predictor predicts the three-dimensional structure using this 1D sequence, and the evaluator independently judges the quality of the predicted results by comparing them to the three-dimensional structure provided by the experimentalist (provided only to the evaluator). CASP conducts true blind reviews and records periodic performance jumps associated with methodological innovation. At the 14th CASP Conference in 2020, AlphaFold’s prediction results showed such a leap in performance that the organizers announced that the 3D structure prediction problem had been solved: the accuracy of most predictions was close to that of experimental measurements.
The broader significance is that Hassabis and Jumper’s work convincingly demonstrates how AI-ML could transform science. Its research shows that AI-ML can build complex scientific hypotheses from multiple data sources, that attention mechanisms (similar to those in ChatGPT) can discover key dependencies and correlations in data sources, and that AI-ML can self-judge the quality of its output results. AI-ML is essentially doing science.
Post time: Sep-23-2023